Chapter 22 Algorithms for Graph Partitioning
22.1 Spectral Clustering
Spectral clustering is a term that data-miners have given to the partitioning problem as it arose in graph theory. The theoretical framework for spectral clustering was laid in 1973 by Miroslav Fiedler [31], [32]. We will begin with a discussion of this early work, and then take a look at how others have adapted the framework to meet the needs of data clustering. In this setting, we have a graph \(G\) on a set of vertices \(N=\{1,2,\dots,n\}\) with edge set \(E=\{(i,j) : i,j \in N \mbox{and} i \leftrightarrow j\}\). Edges between the vertices are recorded in an adjacency matrix \(\A = (a_{ij})\), where \(a_{ij}\) is equal to the weight of the edge connecting vertex (object) \(i\) and vertex \(j\) and \(a_{ij}=0\) if \((i,j) \notin E\). For the immediate discussion, we will assume the graph has no “self-loops,” i.e. \(a_{ii}=0 \forall i\). Spectral clustering algorithms typically involve the Laplacian matrix associated with a graph. A Laplacian matrix is defined as follows:
Definition 22.1 (The Laplacian Matrix) The Laplacian Matrix, \(\mathbf{L}\), of an undirected, weighted graph with adjacency matrix \(\A=(a_{ij})\) and diagonal degree matrix \(\mathbf{D}=\mbox{diag}(\A\e)\) is: \[\mathbf{L}=\mathbf{D}-\A\]
The Laplacian matrix is symmetric, singular, and positive semi-definite. To see this third property, construct an \(n \times |E|\) “vertex-edge incidence” matrix \(\U\) with rows corresponding to vertices and columns corresponding to edges. Allow the edges of the original graph to be directed arbitrarily, and set \[ \U_{v,e} = \left\{ \begin{array}{lr} +\sqrt{a_{ij}} : &\mbox{if } v \mbox{ is the head of } e\\ -\sqrt{a_{ij}} : &\mbox{if } v \mbox{ is the tail of } e\\ 0 : &\mbox{otherwise} \end{array} \right. \]
Then \(\mathbf{L}=\U\U^T\) is positive semi-definite [32]. \(\mathbf{L}\) gives rise to a nice quadratic form:
\[\begin{equation} \tag{22.1} \mathbf{y}^T \mathbf{L} \mathbf{y} = \sum_{(i,j) \in E}a_{ij} (y_i - y_j)^2. \end{equation}\]
Let \(\sigma(\mathbf{L})=\{\lambda_1 \leq \lambda_2 \leq \dots \leq \lambda_n\}\) be the spectrum of \(\mathbf{L}\). Since \(\mathbf{L}\) is positive semi-definite, \(\lambda_i \geq 0 \forall i\). Also, since the row sums of \(\mathbf{L}\) are zero, \(\lambda_1=0\). Furthermore if the graph, \(G\), is composed of \(k\) connected components, each disconnected from each other, then \(\lambda_1=\lambda_2=\dots=\lambda_k = 0\) and \(\lambda_j \geq 0 \mbox{ for } j\geq k+1\). In [32] Fiedler defined the algebraic connectivity of the graph as the second smallest eigenvalue, \(\lambda_2\), because its magnitude provides information about how easily the graph is to be disconnected into two components. Later, in [31], he alluded to the utility of the eigenvector associated with \(\lambda_2\) in determining this two-component decomposition of a graph.
22.2 Fiedler Partitioning
Suppose we wish to decompose our graph into two components (or clusters of vertices) \(C_1\) and \(C_2\) where the edges exist more frequently and with higher weight inside the clusters than between the two clusters. In other words, we intend to make an edge-cut disconnecting the graph into two clusters. It is desired that the resulting partition satisfies the following objectives:- minimize the total weight of edges cut (edges in between components)
- maximize the total weight of edges inside the two components.
To begin with, lets take the quadratic form in Equation ?? and let \(\y\) be a vector that determines the cluster membership of each vertex as follows: \[ \y_i=\left\{ \begin{array}{lr} +1 & : \mbox{if vertex } i \mbox{ belongs in } C_1\\ -1 & : \mbox{if vertex } i \mbox{ belongs in } C_2\\ \end{array} \right. \] Our first goal is then to minimize Equation (22.1) over all such vectors \(\y\):
\[\begin{equation} \tag{22.2} \min_{\y} \y^T \mathbf{L} \y = \sum_{(i,j) \in E} a_{ij} (\y_i-\y_j)^2 = 2 \sum_{\substack{(i,j) \in E \\i \in C_1, j \in C_2}} 4 a_{ij} \end{equation}\]
Note that the final sum is doubled to reflect the fact that each edge connecting \(C_1\) and \(C_2\) will be counted twice. However, the above formulation is incomplete because it does not take into account the second objective, which is to maximize the total weight of edges inside the two components. Indeed it seems the minimum solution to Equation ?? would often involve cutting all of the edges adjacent to a single vertex of minimal degree, disconnecting the graph into components of size \(1\) and \(n-1\), which is generally undesirable. In addition, the above optimization problem is NP-hard. To solve the latter problem, the objective function is relaxed from discrete to continuous. By the Rayleigh theorem, \[\min_{\|\y\|_2=1} \y^T\mathbf{L} \y = \lambda_1\] with \(\y^*\) being the eigenvector corresponding to the smallest eigenvalue. However, for the Laplacian matrix, \(y^*=\e\). In context, this makes sense - in order to minimize the weight of edges cut, we should simply assign all vertices to one cluster, leaving the second empty. In order to divide the vertices into two clusters we need an additional constraint on \(\y\). Since clusters of relatively balanced size are desirable, a natural constraint is \(\y^T\e=0\). By the Courant-Fischer theorem,
\[\begin{equation} \tag{22.3} \min_{\substack{\| \y \|_2=1 \\ \y^T \e=0}} \y^T \mathbf{L} \y = \lambda_2 \end{equation}\]
with \(\y^*=\textbf{v}_2\) being the eigenvector corresponding to the second smallest eigenvalue, \(\lambda_2\). This vector is often referred to as the Fiedler vector after the man who identified its usefulness in graph partitioning. We define the Fiedler graph partition as follows:
Definition 22.2 (Fiedler Graph Partition) Let \(G=(N,E)\) be a connected graph on vertex set \(N=\{1,2,\dots,n\}\) with adjacency matrix \(\A\). Let \(\mathbf{L}=\mathbf{D}-\A\) be the Laplacian matrix of \(G\). Let \(\textbf{v}_2\) be an eigenvector corresponding to the second smallest eigenvalue of \(\mathbf{L}\). The Fiedler partition is:
\[\begin{eqnarray*} C_1 &=& \{i \in N : \textbf{v}_2(i) <0\}\\ C_2 &=& \{i \in N : \textbf{v}_2(i) >0\} \end{eqnarray*}\] Vertices \(j\), for which \(\textbf{v}_2(j)=0\), can be arbitrarily placed into either cluster.
There is no uniform agreement on how to determine the cluster membership of vertices for which \(\textbf{v}_2(j)=0\). The decision to make the assignment arbitrarily comes from experimental results that indicate in some scenarios these zero valuated vertices are equally drawn to either cluster. Situations where there are a large proportion of zero valuated vertices may be indicative of a graph which does not conform well to Fiedler’s partition, and we suggest the user tread lightly in these cases. Figure 22.1 shows the experimental motivation for our arbitrary assignment of zero valuated vertices. The vertices in these graphs are labelled according to the sign of the corresponding entry in \(\textbf{v}_2\). We highlight the red vertex and watch how its sign in \(\textbf{v}_2\) changes as nodes and edges are added to the graph.
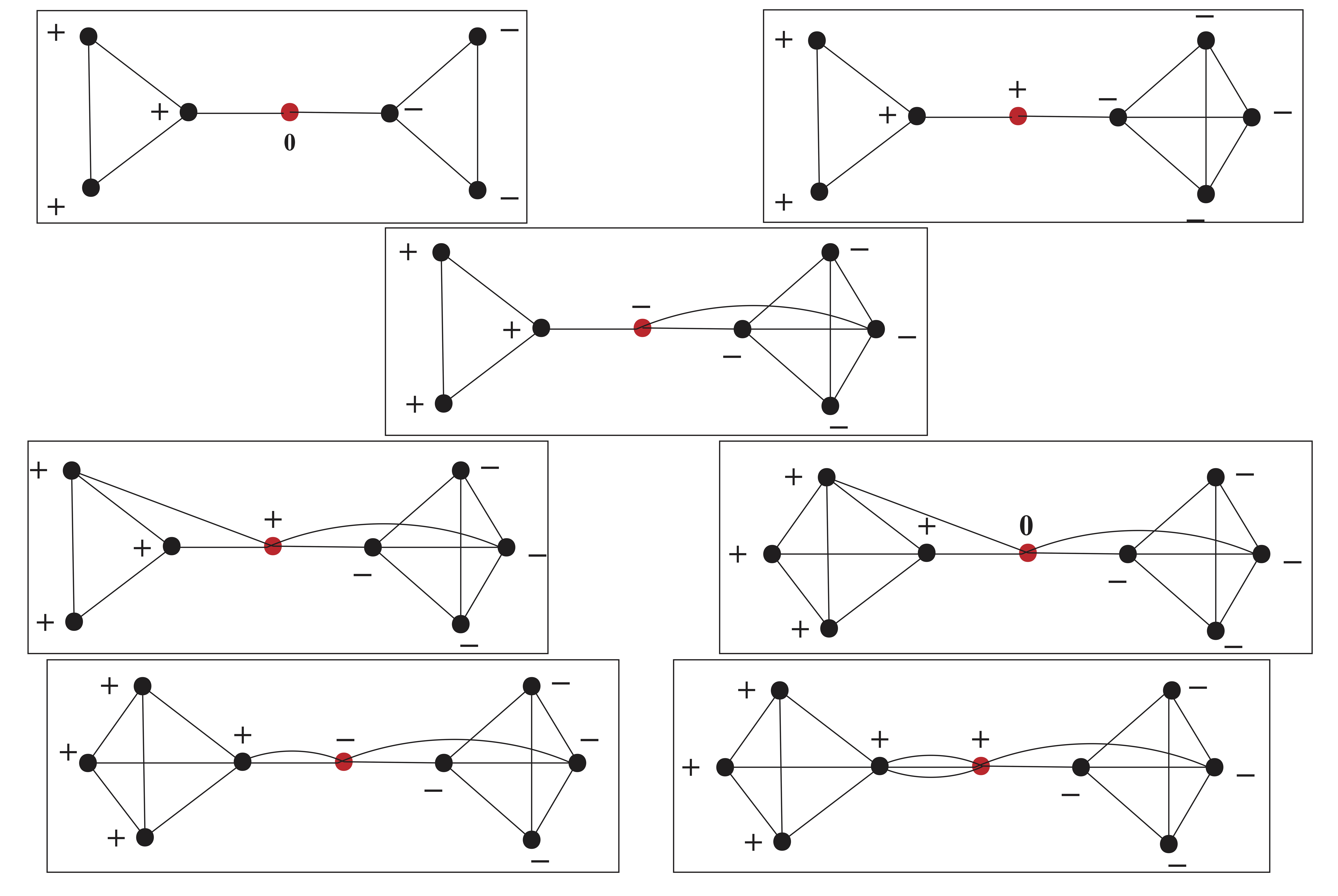
Figure 22.1: Fiedler Partitions and Zero Valuated Vertices
In order to create more than two clusters, the Fiedler graph partition can be performed iteratively, by examining the subgraphs induced by the vertices in \(C_1\) and \(C_2\) and partitioning each based upon their own Fiedler vector, or in an extended fashion using multiple eigenvectors. This iterative method requires a cluster to be chosen for further division, perhaps based upon the algebraic connectivity of the cluster. It is also possible to use the sign patterns in subsequent eigenvectors to further partition the graph. This approach is called Extended Fiedler Clustering and is discussed in Section 22.2.1.1. First, let’s take a more rigorous look at why the sign patterns of the Fiedler vector provide us with a partition.
22.2.1 Linear Algebraic Motivation for the Fiedler vector
The relaxation proposed in Equation (22.3) does not give us a general understanding of why the sign patterns of the Fiedler vector are useful in determining cluster membership information. We will use the following facts:
Lemma 22.1 (Fiedler Lemma 1) Let \(\mathbf{L}=\mathbf{D}-\A\) be a Laplacian matrix for a graph \(G=(V,E)\) with \(|V|=n\). Let \(\sigma(L)=\lambda_1 \leq \lambda_2 \leq \dots \leq \lambda_n\) Then \(\mathbf{L}\) is symmetric and positive semi-definite with \(\lambda_1=0\).
Lemma 22.2 (Fiedler Lemma 2) \(\lambda_2(L)=0\) if and only if the graph \(G\) has 2 components, \(C_1\) and \(C_2\) which are completely disconnected from each other (i.e. there are no edges connecting the vertices in \(C_1\) to the vertices in \(C_2\).)
Lemma 22.3 (Fiedler Lemma 3) Let \(\mathbf{M}\) be a symmetric matrix of rank \(r\). Let \[\mathbf{M} = \mathbf{V} \mathbf{D} \mathbf{V}^T = (\textbf{v}_1, \textbf{v}_2, \dots, \textbf{v}_r) \left( \begin{matrix} \sigma_1 & 0 & \ldots & 0\\ 0 & \sigma_2 & \ldots & 0\\ \vdots & \vdots & \ddots& \vdots\\ 0 & 0 &\ldots & \sigma_r\\ \end{matrix} \right) \left( \begin{matrix} \textbf{v}_1^T\\ \textbf{v}_2^T\\ \vdots \\ \textbf{v}_r^T\\ \end{matrix} \right) =\sum_{i=1}^r \sigma_i \textbf{v}_i \textbf{v}_i^T\] be the singular value decomposition of \(\mathbf{M}\), with \(\sigma_1 \geq \sigma_2 \geq \dots \sigma_r\). Let \(\widetilde{\mathbf{M}}\) be the closest (in the euclidean sense) rank \(k\) approximation to \(\mathbf{M}\): \[\widetilde{\mathbf{M}}=\mbox{arg}\min_{\mathbf{B} , rank(\mathbf{B})=k}\|\mathbf{M} - \bf B\|.\] Then \(\widetilde{\mathbf{M}}\) is given by the truncated singular value decomposition: \[B=\sum_{i=1}^k \sigma_i \textbf{v}_i \textbf{v}_i^T\]
From these simple tools, we can get a sense for why the signs of the Fiedler vector will determine a natural partition of a graph into two components. In the simplest case, \(\lambda_1=\lambda_2=0\), the graph contains two disjoint components, \(C_1\) and \(C_2\). Thus, there exists some permutation matrix \(\mathbf{P}\) such that \(\mathbf{P} \mathbf{L} \mathbf{P}\) is block diagonal:
\[ \mathbf{P} \mathbf{L} \mathbf{P} =\left( \begin{array}{cc} \mathbf{L}_1 & 0\\ 0 & \mathbf{L}_2\\ \end{array} \right) \]
and \(\mathbf{L}_1\) is the Laplacian matrix for the graph of \(C_1\) and \(\mathbf{L}_2\) is the Laplacian for \(C_2\). Let \(n_1\) and \(n_2\) be the number of of vertices in each component. Clearly the eigenvectors \(\textbf{v}_2\) and \(\textbf{v}_2\) associated with \(\lambda_1=0\) and \(\lambda_2=0\) are contained in the span of \(\mathbf{u}_1\) and \(\mathbf{u}_2\) where:
\[\begin{equation} \mathbf{u}_1 = \begin{array}{cc}\begin{array}{c} 1\\ \vdots \\ n_1 \\ n_1+1 \\ \vdots \\ n \end{array} &\left( \begin{array}{c} 1 \\ \vdots \\ 1 \\ 0 \\ \vdots \\ 0 \end{array} \right) \end{array} \qquad\mbox{and}\qquad \mathbf{u}_2 =\begin{array}{cc}\begin{array}{c} 1\\ \vdots \\ n_1 \\ n_1+1 \\ \vdots \\ n \end{array} &\left( \begin{array}{c}0 \\ \vdots \\ 0 \\ 1 \\ \vdots \\ 1 \end{array} \right)\end{array} \end{equation}\]
For all Laplacian matrices, it is convention to consider \(\textbf{v}_1=\e\), the vector of all ones. Under this convention, the two conditions \(\textbf{v}_2 \perp \textbf{v}_1\) and \(\textbf{v}_2 \in span \{ \mathbf{u}_1,\mathbf{u}_2\}\) necessarily force \(\textbf{v}_2\) to have a form such that the component membership of each vertex is discernible from the sign of the corresponding entry in the vector: \[\textbf{v}_2 = \alpha(\mathbf{u}_1-\frac{n_1}{n_2} \mathbf{u}_2).\]
The case when \(\mathbf{L}\) is not disconnected into two components is far more interesting, as this is generally the problem encountered in practice. We will start with a connected graph, so that our Laplacian matrix has rank \(n-1\). Let the spectral decomposition of our Laplacian matrix \(\mathbf{L}\) (which is equivalent to its singular value decomposition because \(\mathbf{L}\) is symmetric) be: \[\mathbf{L}=\sum_{i=2}^n \lambda_i \textbf{v}_i \textbf{v}_i^T.\] Let \(\widetilde{\mathbf{L}}\) be the closest rank \(n-2\) approximation to \(\mathbf{L}\). Then, by Lemmas 22.2 and 22.3, \[\widetilde{\mathbf{L}} = \sum_{i=3}^n \lambda_i \textbf{v}_i \textbf{v}_i^T\] and there exists a permutation matrix \(\mathbf{P}\) such that
\[ \mathbf{P} \widetilde{\mathbf{L}} \mathbf{P} =\left( \begin{matrix} \widetilde{\mathbf{L}}_1 & 0\\ 0 & \widetilde{\mathbf{L}}_2 \end{matrix} \right). \]
Suppose we permute the rows of \(\mathbf{L}\) accordingly so that
\[ \mathbf{P} \mathbf{L} \mathbf{P} =\left( \begin{matrix} \mathbf{L}_1 & -\mathbf{E}\\ -\mathbf{E}^T & \mathbf{L}_2\\ \end{matrix} \right) \] where \(\mathbf{E}_{ij} \geq 0 \forall i,j\) because \(\mathbf{L}\) is a Laplacian matrix. Consider the difference between \(\mathbf{L}\) and \(\widetilde{\mathbf{L}}\):
\[\mathbf{L}-\widetilde{\mathbf{L}} = \lambda_2 \textbf{v}_2 \textbf{v}_2^T\]
which entails \(\mathbf{L} - \lambda_2 \textbf{v}_2 \textbf{v}_2^T = \widetilde{\mathbf{L}}\). If we permute the vector \(\textbf{v}_2\) in the same manner as the matrices \(\mathbf{L}\) and \(\widetilde{\mathbf{L}}\), then one thing is clear:
\[\begin{equation*} \lambda_2\mathbf{P}\textbf{v}\textbf{v}_2^T\mathbf{P} = \left(\begin{matrix} \A & -\mathbf{E} \\ -\mathbf{E}^T & B \end{matrix}\right) \end{equation*}\] Thus, if \[\begin{equation*}\mathbf{P}\textbf{v} =\left(\begin{matrix} \mathbf{a} \\ \mathbf{b}\\ \end{matrix} \right) \end{equation*}\]
where \(\mathbf{a} \in \Re^{n_1}\) and \(\mathbf{b} \in \Re^{n_2}\)
22.2.1.1 Extended Fiedler Clustering
In the extended Fiedler algorithm, we use the sign patterns of entries in the first \(l\) eigenvectors of \(\mathbf{L}\) to create up to \(k=2^l\) clusters. For instance, suppose we had 10 vertices, and used the \(l=3\) eigenvectors \(\textbf{v}_2,\textbf{v}_3,\mbox{ and }\textbf{v}_4\). Suppose the sign of the entries in these eigenvectors are recorded as follows: \[ \begin{array}{cc} & \begin{array}{ccc} \mathbf{v}_2 & \mathbf{v}_3&\mathbf{v}_4 \end{array}\cr \begin{array}{c} 1 \\ 2 \\ 3 \\ 4 \\ 5 \\ 6 \\ 7 \\ 8 \\ 9 \\ 10 \end{array} & \left( \begin{array}{ccc} +&+&-\cr -&+&+\cr +&+&+\cr -&-&-\cr -&-&-\cr +&+&-\cr -&-&-\cr -&+&+\cr +&-&+\cr +&+&+ \end{array} \right) \end{array} \] Then the 10 vertices are clustered as follows: \[ \{1,6\},\quad \{2,8\},\quad \{3,10\},\quad \{4,5,7\},\quad \{9\}. \] Extended Fiedler makes clustering the data into a specified number of clusters \(k\) difficult, but may be able determine a natural choice for \(k\) as it partitions the data along several eigenvectors.
In a 1990 paper by Pothen, Simon and Liou, an alternative formulation of the Fiedler partition is proposed [26]. Rather than partition the vertices based upon the sign of their corresponding entries in \(\mathbf{v}_2\), the vector \(\mathbf{v}_2\) is instead divided at its median value. The main motivation for this approach was to split the vertices into sets of equal size. In 2003, Ding et al. derived an objective function for determining an ideal split point for similar partitions using the second eigenvector of the normalized Laplacian, defined in Section 22.2.2.2 [27]. The basic idea outlined above has been adapted and altered hundreds if not thousands of times in the past 20 years. The present discussion is meant merely as an introduction to the literature.
22.2.2 Graph Cuts
The majority of spectral algorithms are derived as alterations of the objective function in Equation (22.3).The idea is the same: partition the graph into two components by means of a minimized edge cut, while requiring that the two components remain somewhat balanced in size (i.e. do not simply isolate a small number of vertices). Two common objective functions which embody this idea are the ratio cut (RatioCut) [24], the normalized cut (Ncut) [57].
22.2.2.1 Ratio Cut
The ratio cut objective function was first introduced by Hagen and Kahng in 1992 [24]. Given a graph \(G(V,E)\) with vertex set \(V\) partitioned into \(k\) disjoint clusters, \(V_1,V_2,\dots V_k\), the ratio cut of the given partition is defined as \[\mbox{RatioCut}(V_1,V_2,\dots,V_k) = \sum_{i=1}^k \frac{w(V_i,\bar{V_i})}{|V_i|}\] where \(|V_i|\) is the number of vertices in \(V_i\), \(\bar{V_i}\) is the complement of the set \(V_i\) and, given two vertex sets \(A\) and \(B\), \(w(A,B)\) is the sum of the weights of the edges between vertices in \(A\) and vertices in \(B\). Let \(\mathbf{H}\) be an \(n\times k\) matrix indicating cluster membership of vertices by its entries: \[\begin{equation} \tag{22.4} \mathbf{H}_{ij}= \begin{cases} \frac{1}{\sqrt{|V_j|}}, & \mbox{if the } i^{th} \mbox{ vertex is in cluster } V_j \\ 0 & \text{otherwise} \end{cases} \end{equation}\]
Then \(\mathbf{H}^T\mathbf{H}=\mathbf{I}\) and minimizing the ratio cut over all possible partitionings is equivalent to minimizing \[f(\mathbf{H}) = \mbox{Trace}(\mathbf{H}^T\mathbf{L}\mathbf{H})\] over all matrices \(\mathbf{H}\) described by Equation (22.4), where \(\mathbf{L}\) is the Laplacian matrix from Definition 22.1. The exact minimization of this objective function is again NP-hard, but relaxing the conditions on \(\mathbf{H}\) to \(\mathbf{H}^T\mathbf{H}=\mathbf{I}\) yields a solution \(\mathbf{H}^*\) with columns containing the eigenvectors of \(\mathbf{L}\) corresponding to the \(k\) smallest eigenvalues.
Unfortunately, after this relaxation it is not necessarily possible to automatically determine from \(\mathbf{H}^*\) which vertices belong to each cluster. Instead, it is necessary to look for clustering patterns in the rows of \(\mathbf{H}^*\). This is a common conceptual drawback of the relaxation of objective functions in spectral clustering. The best way to procede after the relaxation is to cluster the rows of \(\mathbf{H}^*\) with an algorithm like \(k\)-means to determine a final clustering. The ratio cut minimization method is generally referred to as unnormalized spectral clustering [72]. The algorithm is as follows:
Input: \(n \times n\) adjacency (or similarity) matrix \(\mathbf{A}\) for a graph on vertices (or objects) \(\{1,\dots,n\}\) and desired number of clusters \(k\)
|
22.2.2.2 Normalized Cut (Ncut)
The normalized cut objective function was introduced by Shi and Malik in 2000 [57]. Given a graph \(G(V,E)\) with vertex set \(V\) partitioned into \(k\) disjoint clusters, \(V_1,V_2,\dots V_k\), the normalized cut of the given partition is defined as \[\mbox{Ncut}(V_1,V_2,\dots,V_k)= \sum_{i=1}^k \frac{w(V_i,\bar{V_i})}{\mbox{vol}(V_i)},\] where \(\mbox{vol}(V_i)\) is the sum of the weights of the edges connecting the vertices in \(V_i\). Whereas the size of a subgraph \(V_i\) in the ratio cut formulation is measured by the number of vertices \(|V_i|\), in the normalized cut formulation it is measured by the total weight of the edges in the subgraph. Thus, minimizing the normalized cut is equivalent to minimizing \[f(\mathbf{H}) = \mbox{Trace}(\mathbf{H}^T\mathbf{L}\mathbf{H})\] over all matrices \(\mathbf{H}\) with the following form:
\[\begin{equation} \mathbf{H}_{ij}= \begin{cases} \frac{1}{\sqrt{\mbox{vol}(V_j)}}, & \mbox{if the } i^{th} \mbox{ vertex is in cluster } V_j \\ 0 & \text{otherwise.} \end{cases} \tag{22.5} \end{equation}\]
With \(\mathbf{H}^T \mathbf{D} \mathbf{H} = \mathbf{I}\) where \(\mathbf{D}\) is the diagonal degree matrix from Definition 22.1. Thus, to relax the problem, we substitute \(\mathbf{G}=\mathbf{D}^{1/2}\mathbf{H}\) and minimize \[f(\mathbf{G})=\mathbf{G}^T \mathscr{L} \mathbf{G}\] subject to \(\mathbf{G}^T\mathbf{G}=\mathbf{I}\), where \(\mathscr{L}=\mathbf{D}^{-1/2}\mathbf{L} \mathbf{D}^{-1/2}\) is called the normalized Laplacian. Similarly, the solution to the relaxed problem is the matrix \(\mathbf{G}^*\) with columns containing eigenvectors associated with the \(k\) smallest eigenvalues of \(\mathscr{L}\). Again, the immediate interpretation of the entries in \(\mathbf{G}^*\) is lost in the relaxation and so a clustering algorithm like \(k\)-means is used to determine the patterns.
Input: \(n \times n\) adjacency (or similarity) matrix \(\mathbf{A}\) for a graph on vertices (or objects) \(\{1,\dots,n\}\) and desired number of clusters \(k\)
|
22.2.2.3 Other Normalized Cuts
While the algorithm in Table 22.2 carries “normalized cut” in its title, other researchers have suggested alternative ways to consider normalized cuts in a graph. In a popular 2001 paper, Ng, Jordan, and Weiss made a slight alteration of the previous algorithm which simply normalized the rows of the eigenvector matrix computed in step 2 to have unit length before proceeding to step 3 [58]. This algorithm is presented in Table 22.3.
Input: \(n \times n\) adjacency (or similarity) matrix \(\mathbf{A}\) for a graph on vertices (or objects) \(\{1,\dots,n\}\) and desired number of clusters \(k\)
|
In 2001, Meila and Shi altered the objective function once again, and derived yet another spectral algorithm using the normalized random walk Laplacian, \(\mathscr{L}_{rw} = \mathbf{D}^{-1}\mathbf{L} = \mathbf{I} - \mathbf{D}^{-1} \A\) [64]. As shown in [52], if \(\lambda\) is an eigenvalue for \(\mathscr{L}\) with corresponding eigenvector \(\mathbf{v}\) then \(\lambda\) is also an eigenvalue for \(\mathscr{L}_{rw}\) with corresponding eigenvector \(\mathbf{D}^{1/2}\mathbf{v}\). This formulation amounts to a different scaling of the eigenvectors in step 3 of Table 22.3. This normalized random walk Laplacian will present itself again in Section 22.2.3. Meila and Shi’s spectral clustering method is outlined in Table 22.4.
Input: \(n \times n\) adjacency (or similarity) matrix \(\mathbf{A}\) for a graph on vertices (or objects) \(\{1,\dots,n\}\) and desired number of clusters \(k\)
Output: Clusters \(C_1,\dots,C_k\) such that \(C_j = \{i : \mathbf{y}_i \in \bar{C}_j\|\) |
All of the spectral algorithms outlined thus far seem very similar in their formulation, yet in practice they tend to produce quite different results. This presents a problem because while each method has merit in its own right, it is impossible to predict which one will work best on any particular graph. We will discuss this problem further in .
22.2.3 Power Iteration Clustering
In a 2010 paper, Frank Lin and William Cohen propose a fast, scalable algorithm for clustering graphs using the power method (or power iteration) [71]. Let \(\mathbf{W}=\mathbf{D}^{-1}\A\) be the \(n\times n\) row-normalized (row stochastic) adjacency matrix for a graph, and let \(\mathbf{v}_0 \neq 0\) be a vector in \(\Re^n\). A simple method for computing the eigenvector corresponding to the largest eigenvalue of \(\mathbf{W}\) is the power method, which repeatedly computes the power iteration \[\mathbf{v}_{t+1}=c\mathbf{W}\mathbf{v}_t\] where \(c=1/\|\mathbf{W}\mathbf{v}_t\|_1\) is a normalizing constant to prevent \(\mathbf{v}_t\) from growing too large.
Applying the power method to convergence on \(\mathbf{W}\) would result in the uniform vector \(\alpha \e\) where \(\alpha = 1/n.\) However, stepping through a small number of power iterations, will result in a vector that contains combined information from the eigenvectors associated with the largest eigenvalues. The formulation of Meila and Shi’s spectral algorithm in [64] warranted the use of the eigenvectors corresponding to the \(k\) smallest eigenvalues of the normalized random walk Laplacian \(\mathscr{L}_{rw} = \mathbf{I}-\mathbf{W}\) which is equivalent to the consideration of the eigenvectors of the largest eigenvalues of \(\mathbf{W}\). Thus, the idea behind Power Iteration Clustering (PIC) is to detect and stop the power method at some number of iterations \(t\) such that \(\mathbf{v}_t\) is a useful linear combination of the first \(k\) eigenvectors. The analysis in [71] motivates the idea that the power method should pass through some initial stage of local convergence at the cluster level before going on to the stage of global convergence toward the uniform vector. At this stopping point, it is expected that \(\mathbf{v}_t\) will be an approximately piecewise constant vector, nearly uniform on each of the clusters. Thus, the clusters at this stage will be revealed by the closeness of their corresponding entries in \(\mathbf{v}_t\). See [71] for the complete analysis. The PIC procedure is given in Table 22.5.
Applying the power method to \(\mathbf{P}^T\) would equate to watching the probability distribution of a random walker evolve through time steps of the Markov Chain, where \(\mathbf{v}_t\) is the distribution at time \(t\), and eventually would converge to the stationary distribution in Equation (22.10).
However, according to Lin and Cohen, stepping through a limited number of power iterations on \(\mathbf{P}\) is equivalent to observing the same chain backwards, so that \(\mathbf{v}_t(i)\) gives the observer a sense of the most likely distribution of the chain \(t\) steps in the past, given that the walk ended with distribution \(\mathbf{v}_0\). On a graph with cluster structure as described above, a random walker that ends up on a particular vertex \(j \in C_1\) is more or less equally likely to have come from any other node in \(C_1\) (but relatively unlikely to have come from \(C_i, i\neq1\)), making the distribution close to uniform on the vertices in \(C_1\). The same argument is true for any cluster \(C_j\) and thus, by stepping backwards through time we expect to find these distribution vectors which are nearly uniform on each cluster \(C_j\). For a complete discussion of the algorithm, including a more detailed mathematical analysis, consult [71].
Input: A row-stochastic matrix \(\mathbf{P}=\mathbf{D}^{-1}\A\) where \(\A\) is an adjacency or similarity matrix and the number of clusters \(k\).
Output: Clusters \(C_1, C_2,\dots,C_k\). |
22.2.4 Clustering via Modularity Maximization
Another technique proposed in the network community detection literature compares the structure of a given graph to what one may expect from a random graph on the same vertices [29], [30]. The motivation for this method was that simply counting edges between clusters as was done in previous spectral methods may not be the best way to define clusters in graph. A better approach may be to somehow measure whether they are fewer edges than expected between communities. Let \(\A\) be the adjacency matrix of the graph (or network) and let \(\mathbf{P}\) be the adjacency matrix of a random graph on the same vertices containing the expected value of weights on that graph. Then the matrix \(\mathbf{B}=\A-\mathbf{P}\) would contain information about how the structure of \(\A\) deviates from what is expected. Obviously this formulation relies on some underlying probability distribution of the weights in the random graph, known as the null model. The most common null model uses the degree sequence of the vertices in the given graph, \(\{d_1,d_2,\dots,d_n\}\), where \(d_i\) is the degree of vertex \(i\) (i.e. \(d_i\) is the sum of the weights of the edges connected to vertex \(i\)), to create the probabilities [30] \[\begin{equation} \tag{22.6} p(\mbox{edge}(i,j)) = \frac{d_j}{\sum_{k=1}^n d_k}. \end{equation}\]
Thus, the expected value of the weight of the edge from \(i\) to \(j\) is \[\mathbf{P}_{ij} = E(w(i,j)) = d_i \left(\frac{d_j}{\sum_{k=1}^n d_k}\right).\]
One may recognize that the probabilities in Equation (22.6) are precisely the stationary probabilities of the random walk on the graph defined by \(\A\), and thus seem a reasonable choice for a null model. This formulation gives us \(E(w(i,j)) = E(w(j,i))\) as desired for an undirected graph. Using this null model, a modularity matrix \(\mathbf{B}\) is formed as \[\mathbf{B} = \A - \mathbf{P}.\]
For a division of the data into two clusters, let \(\mathbf{s}\) be an \(n\times 1\) vector indicating cluster membership by \[\mathbf{s}_{i} = \begin{cases} -1 &: \mbox{vertex } i \mbox{ belongs in cluster 1}\\ \,\,\,\,1 &: \mbox{vertex } i \mbox{ belongs in cluster 2} \end{cases}. \] Let \(d=\sum_{k=1}^n d_k\). The modularity of a given partition is defined by \[\begin{equation} \tag{22.7} Q= \frac{1}{2d} \sum_{i,j} \mathbf{B}_{ij} \mathbf{s}_i\mathbf{s}_j = \frac{1}{2d}\mathbf{s}^T\mathbf{B}\mathbf{s}. \end{equation}\] The goal of the algorithm proposed in [30] is to maximize this quantity, thus we can drop the constant \(1/2d\) and write the objective as \[\begin{equation} \tag{22.8} \max_{\substack{\mathbf{s} \\ \mathbf{s}_i = \pm 1}} Q = \mathbf{s}^T \mathbf{B} \mathbf{s} \end{equation}\]
Illustrative Example
To get an idea of why this is true, consider the case where we have two relatively obvious clusters \(C_1\) and \(C_2\) in a graph and reorder the rows and columns of the adjacency matrix to reflect this structure, \[ \A=\left[ \begin{array}{cc} \A_{C_1} & \mathbf{E} \\ \mathbf{E}^T & \A_{C_2} \end{array}\right] \] Where \(\A_{C_1}\) and \(\A_{C_2}\) are relatively dense matrices with larger entries representing the weight of edges within the clusters \(C_1\) and \(C_2\) respectively and \(\mathbf{E}\) is a sparse matrix with smaller entries representing the weight of edges which connect the two clusters. In a random graph with no community or cluster structure, we’d be likely to find just as many edges between the clusters as within clusters. Thus, after subtracting \(\mathbf{P}\) our modularity matrix may look something like \[ \mathbf{B}=\left[ \begin{array}{cc} \mathbf{B}_{11} & \mathbf{B}_{12} \\ \mathbf{B}_{21} & \mathbf{B}_{22} \end{array}\right] \approx\left[ \begin{array}{cc} + & - \\ - & + \end{array}\right] \] Where the indicated signs reflect the sign tendancy of values in \(\mathbf{B}\). In other words, the entries in the diagonal blocks \(\mathbf{B}_{11}\) and \(\mathbf{B}_{22}\) tend to be positive because the edges within clusters had larger weights than one would expect at random and the entries in the off diagonal blocks \(\mathbf{B}_{12}\) and \(\mathbf{B}_{21}\) tend to be negative because the edges between clusters had smaller weights than one would expect at random. Thus, the modularity of this graph, \(\mathbf{Q} = \mathbf{s}^T \mathbf{B} \mathbf{s}\), will be maximized by the appropriate partition \(\mathbf{s}^T=[\mathbf{s}^T_1,\mathbf{s}^T_2]=[\e^T_{C_1}, -\e^T_{C_2}]\).
In order to maximize the modularity objective function given in Equation (22.8), let \(\uu_1,\uu_2,\dots,\uu_n\) be an orthonormal set of eigenvectors for \(\mathbf{B}\) corresponding respectively to the eigenvalues \(\lambda_1 \geq \lambda_2 \geq \dots \geq \lambda_n\). Write the vector \(\mathbf{s}\) as a linear combination of eigenvectors, \[\mathbf{s} = \sum_{i=1}^n \alpha_i \uu_i\] where \[\alpha_i =\uu_i^T \mathbf{s}.\] Then, the objective function from Equation (22.8) becomes \[\max_{\substack{\mathbf{s} \\ \mathbf{s}_i = \pm 1}} \left(\sum_{i=1}^n \alpha_i \uu_i^T \mathbf{B}\right)\left(\sum_{i=1}^n \alpha_i \uu_i\right) = \sum_{i=1}^n \lambda_i (\uu_i^T \mathbf{S})^2.\]
This optimization is NP-hard due to the constraint that \(\mathbf{s}_i =\pm 1\). It is clear that without this constraint one would choose \(\mathbf{s}\) proportional to \(\uu_1\), maximizing the first term in the summation (associated with the largest eigenvalue) and terminating the others. A reasonable way to proceed in light of this information is to maximize the leading term and ignore the remaining terms. To accomplish this, it is quite clear that we should choose \(\mathbf{s}\) so that its entries match the signs of the entries in \(\uu_1\). The placement of vertices corresponding to zero entries in \(\uu_1\) will be decided arbitrarily. However, if the leading eigenvalue of the modularity matrix is negative then the corresponding eigenvector is \(\e\), leading us to no partition. According to [30] the ``no partition’’ solution in this scenario is in fact the correct result, i.e. a negative leading eigenvalue indicates there is no community or cluster structure in the graph. This gives a clear stopping point for the procedure, which allows it to automatically determine an appropriate number of clusters or communities to create. Unfortunately, the arbitrary placement of vertices corresponding to zero entries in \(\uu_1\) may in some cases affect the determined number of clusters.
To create more than 2 clusters, the above procedure can be repeated on each of the subgraphs induced by the vertices in each cluster found. This leads us to an iterative divisive (hierarchical) algorithm like the iterative Fiedler method in Section 22.2.1.1 and PDDP in Section ??. The modularity clustering procedure is formalized in Table .
Input: \(n \times n\) adjacency matrix \(\A\) for an undirected graph to be partitioned
|
22.3 Stochastic Clustering
An alternative way to interpret a graph is by considering a random walk along the edges. For an undirected graph with adjacency matrix \(\A\), we can create a transition probability matrix \(\mathbf{P}\) by dividing each row by the corresponding row sum. Using the degree matrix from Definition 22.1 we have \[\mathbf{P}=\mathbf{D}^{-1}\A.\] If our graph does indeed have some cluster structure, i.e. sets of vertices \(C_1,C_2, \dots,C_k\) for which the total weight of edges within each set are substantially higher than the total weight of edges between the different sets, then a random walker in a given cluster \(C_i\) is more likely to stay in \(C_i\) for several steps than he is to transition to another cluster \(C_j\). It is well known that for a connected and undirected graph, the long term probability distribution is given by \[\begin{equation} \tag{22.10} \pi^T = \frac{\e^T\mathbf{D}}{\e^T\mathbf{D}\e^T} \end{equation}\] Which is not likely to give any cluster information. However, the short-term evolution of this walk can tell us something about the cluster structure because a random walker is far more likely, in the short-run, to remain inside a cluster than he is to transition between clusters. The Stochastic Clustering Algorithm (SCA) of Wessell and Meyer [65] takes advantage of this fact.
22.3.1 Stochastic Clustering Algorithm (SCA)
In a 2012 paper, Chuck Wessell and Carl Meyer formulated a clustering model by creating a symmetric (doubly stochastic) transition matrix \(\mathbf{P}\) [65], [66] from the adjacency matrix of a graph. The method in this paper is quite similar to that in PIC except that here the mathematics of the ``backward’’ Markov Chain intuition given in [71] works out in this context because the probability transition matrix is symmetric. One added feature in this algorithm is the automatic determination of the number of clusters in the data, using eigenvalues of the transition matrix \(\mathbf{P}\). Wessell and Meyer’s formulation is based on theory that was developed by Nobel Laureate economist Herbert Simon and his student Albert Ando. This theory surrounds the mixing rates of resources or wealth in local economies (composed of states in a Markov chain) as part of a global economy (which links together some states from each local economy). It is assumed that the adjacency matrix for the graph is irreducible, or equivalently that the graph is connected.
The basic idea is that resources will be exchanged more frequently at a local level than they will at the global level. Suppose individual companies from a global economy are represented as nodes in a graph with edges between them signifying the amount of trade between each pair of companies. Natural clusters would form in this graph at a local level, represented by the strong and frequent trade relationships of proximal companies. Let \(k\) be the number of local economies (clusters), each containing \(n_i\) states \(i=1,\dots,k\), and define the distribution of resources at time \(t\) as \(\mathbf{\pi}_t\), given a starting distribution \(\mathbf{\pi}_0\). Then \[\mathbf{\pi}_t^T = \mathbf{\pi}_0^T\mathbf{P}^t\]
The heavily localized trade in this global economy leads to a so-called short-term stabilization of the system characterized by a distribution vector at some time \(t\) which is nearly constant across each local economy: \[\mathbf{\pi}_t^T \approx \left( \frac{\alpha_1}{n_1} \frac{\alpha_1}{n_1} \dots \frac{\alpha_1}{n_1} | \frac{\alpha_2}{n_2} \frac{\alpha_2}{n_2} \dots \frac{\alpha_2}{n_2} | \dots | \frac{\alpha_k}{n_k} \frac{\alpha_k}{n_k} \dots \frac{\alpha_k}{n_k} \right)\] After this short-term stabilization, the distribution of goods in the Markov Chain is eventually expected to converge to a constant level across every state. However, in the period following the short-run stabilization, the distribution vector retains its approximately piecewise constant structure for a some time before settling down into its final uniform equilibrium.
Wessell and Meyer’s derivation requires the creation of a symmetric probability transition matrix \(\mathbf{P}\) from the adjacency matrix \(\A\) by means of a simultaneous row and column scaling. In other words, a diagonal matrix \(\mathbf{S}\) is determined for which \[\mathbf{S}\A\mathbf{S}= \mathbf{P}\] is a doubly stochastic transition probability matrix. This task turns out to be quite simple, \(\mathbf{S}\) is found by iterating a single step until convergence. Letting \(\mathbf{S}_{ii}=\mathbf{s}(i)\), the diagonal scaling procedure put forth by Ruiz [[17]} is simply: \[\begin{equation} \tag{22.11} \begin{split} \mathbf{s}_0 &= \e \\ \mathbf{s}_{t+1}(i) &=\sqrt{\frac{\mathbf{s}_t(i)}{\A_{i*}^T\mathbf{s}_t}} \end{split} \end{equation}\]
In [66], it is convincingly argued that the diagonal scaling procedure does not change the underlying cluster structure of the data in \(\A\), and thus that the desired information is not damaged by this transformation. The clusters in this method are found in a similar manner to PIC, where \(k\)-means is employed to find the nearly piecewise constant segments of the distribution vector \(\mathbf{\pi}_t\) after a short number of steps. The Stochastic Clustering Algorithm automatically determines the number of clusters in the data by counting the number of eigenvalues whose value is close to \(\lambda_1=1\). This group of eigenvalues near 1 is referred to as the Perron cluster. We postpone discussion of this matter to where it will be analyzed in detail. For now, we present the Stochastic Clustering method in Table 22.7. The eigenvector iterations in SCA are quite similar to those put forth in PIC, and users commonly create visualizations of the iterations that look quite similar to those in Figure ??.
Input: Adjacency matrix \(\A\) for some graph to be partitioned
|